Precision medicine, also known as personalized medicine, is a key clinical goal for the effective treatment of heterogeneous, complex diseases such as inflammatory bowel disease (IBD), cancer, autoimmune diseases and COVID-19.
Recent paper published in the journal Nature Communications describes a precision medicine approach - the integrated SNP (Single Nucleotide Polymorphism) Network Pipeline (iSNP).
The iSNP tool will help to identify subtype of IBD for every patient based on their specific genetics. It could help to describe the individual pathogenesis story and find the best treatment.
Patients with Inflammatory Bowel Disease (IBD) develop the condition due to distinct and different mechanisms, determined by their genetics. The causes of IBD aren't understood but are linked to dysfunction of the immune system and how it reacts to food and the gut microbiome, including virome.
For IBD, less than 10% of the identified SNPs are in coding regions of genes and over 90% of SNPs are in areas once thought to just be junk DNA, controlling and regulating the activity of the genes. The immune system functions by taking a wide range of different inputs that trigger different signaling networks within the cell, integrating these to produce a balanced, appropriate response, so a combination of even subtlest SNPs could disequilibrate the system. Understanding how they combine to influence intricately interlinked signals would fill in major gaps enabling personalized treatment.
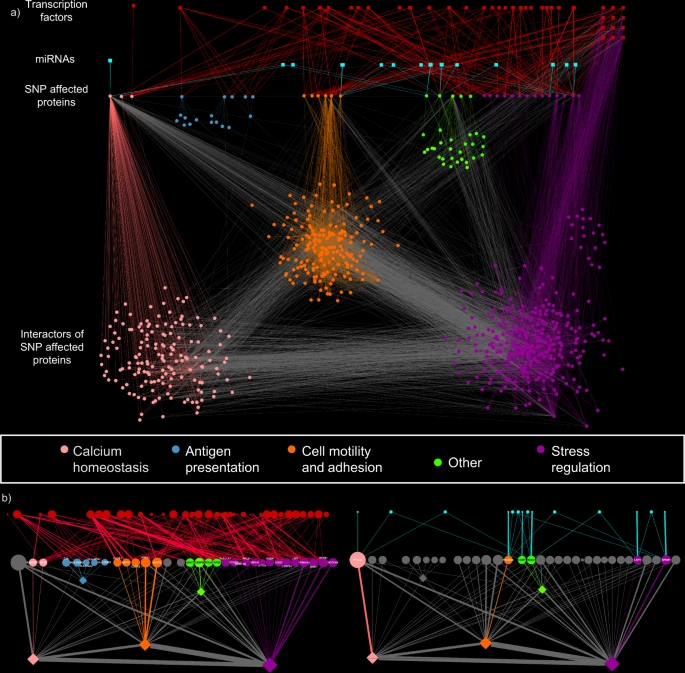
The iSNP workflow identifies patient clusters with distinct pathomechanisms.
Patient data is layered with population-wide genomics and transcriptomics using. To achieve this, hidden proteins contributing to pathogenesis and key pathogenic pathways are identified and aligned with pathological processes in disease development.High-quality individual patient genetic information was used along with preprocessed and quality-controlled immunochip data. miRNA-TS identification algorithm MIRANDA was included in the pipeline along with other genetic analysis tools. A computer simulation of interactions, pathways and networks used databases of known and predicted interactions between proteins in the network.
There was not enough granularity in the clinical data to link all pathways with phenotypes and remove confounders such as recurrent corticosteroid therapy. Further work will need to be done on larger cohorts and with multi-omics datasets to confirm the potential for iSNP to be used for precision therapy based on patient-specific genetics.
REFERENCE
Johanne Brooks-Warburton et al, A systems genomics approach to uncover patient-specific pathogenic pathways and proteins in ulcerative colitis, Nature Communications (2022). DOI: 10.1038/s41467-022-29998-8